Unlocking the Structure of Biofilms
Unlocking the Structure of Biofilms https://pediatricsnationwide.org/wp-content/uploads/2020/04/biofilm-header-1024x575.jpg 1024 575 Kevin Mayhood Kevin Mayhood https://secure.gravatar.com/avatar/bd57a8b155725b653da0c499ae1bf402?s=96&d=mm&r=g- April 10, 2020
- Kevin Mayhood
Researchers characterize a component that stabilizes biofilms, a step toward learning ways to disrupt protection of harmful bacteria.
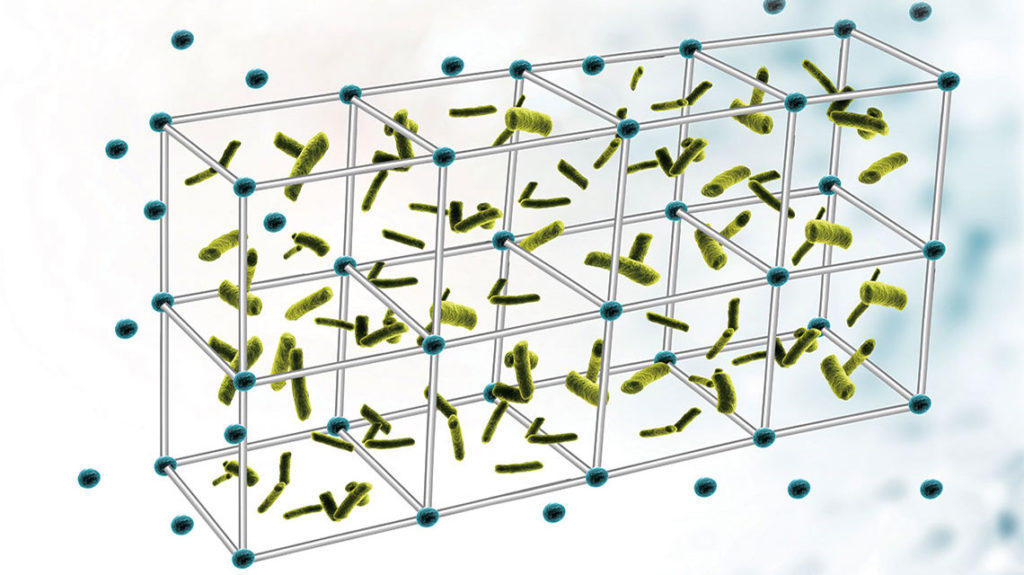
In the extracellular DNA lattice of bacterial biofilms, nature appears to reprise the functional equivalent of Holliday junction (HJ) intermediates — cross-shaped structures formed during the process of genetic recombination, researchers at Nationwide Children’s Hospital report in Proceedings of the National Academy of Sciences.
The HJ-like structures are key to a stable biofilm that protects bacteria. The protection, which includes the biofilm’s resistance to DNA-cleaving nucleases, allows the pathogens to act as a reservoir for chronic and recurrent infections, costing thousands of lives and billions of dollars in care annually.
“Bacteria in a biofilm state are 1,000-fold more resistant to antibiotics than when they are free-living,” says Steven Goodman, PhD, a principal investigator in the Center for Microbial Pathogenesis at Nationwide Children’s and senior author of the research. “There’s plenty of use for preventing biofilm formation, but for practical reasons, getting rid of an extant matrix has much more utility. Usually you don’t have a medical problem until you come to the doctor with biofilm infections.”
Dr. Goodman and colleagues are already developing a potential antibody-based therapy to disrupt biofilms but, “once we understand what the structure is, we’ll be able to find other ways to undermine it. This is a big step in a series of steps to figure that out.”
The research builds on a discovery Dr. Goodman and Lauren Bakaletz, PhD, director of the Center for Microbial Pathogenesis, made nearly a decade ago. The two identified the protein DNABII as a linchpin contributing to the structural integrity of single-species and multispecies biofilms found on devices and in chronic infections of the ear, lung, urinary tract and more.
Due to the HJ-like shape of the lattice structures and DNABII’s affinity and specificity to HJs, the researchers suggested the structures were related to HJs. Aishwarya Devaraj, PhD, a senior research scientist in Dr. Goodman’s lab, tested the hypothesis by establishing equivalence.
On biofilms of uropathogenic Escherichia coli, nontypeable Haemophilus influenzae and Staphylococcus epidermidis, Dr. Devaraj added an antibody the researchers have shown removes DNABII and collapses the biofilm. At the same time, she added the protein RuvA, which binds to and stabilizes HJs.
The structures held and were as strong as before.
She then added the proteins RuvB and RuvC, which when combined with the RuvA, chop HJs and resolve them into two double-stranded DNA helices.
The biofilms collapsed.
“The only reason the RuvABC complex was able to chew up the DNA matrix is we knocked off the DNABII protein,” Dr. Goodman says. “But how these scaffolds form, what the big thick strands of fibers are, why other proteins that degrade DNA have no effect — those are things we’re currently studying.”
Reference:
Devaraj A, Buzzo JR, Mashburn-Warren L, Gloag ES, Novotny LA, Stoodley P, Bakaletz LO, Goodman SD. The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday Junction recombination intermediates. Proceedings of the National Academy of Sciences. 2019 Dec 10;116(50):25068-25077.
Image credit: Nationwide Children’s
About the author
-
Kevin Mayhoodhttps://pediatricsnationwide.org/author/kevin-mayhood/
-
Kevin Mayhoodhttps://pediatricsnationwide.org/author/kevin-mayhood/April 25, 2015
-
Kevin Mayhoodhttps://pediatricsnationwide.org/author/kevin-mayhood/April 25, 2015
-
Kevin Mayhoodhttps://pediatricsnationwide.org/author/kevin-mayhood/April 25, 2015